
|
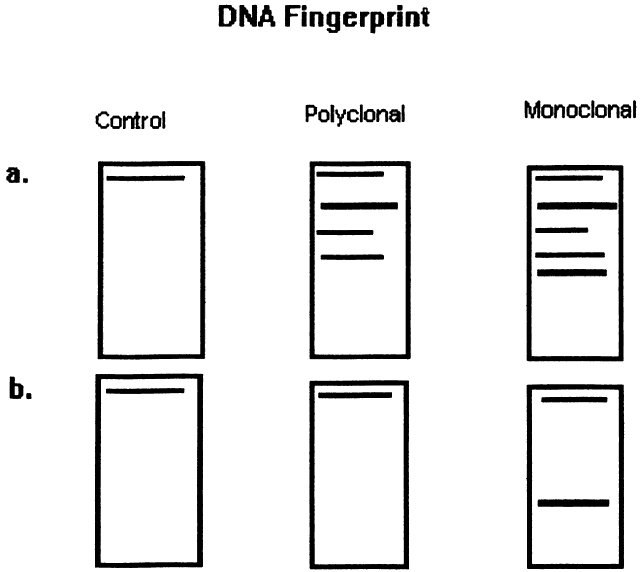
| Fig. 3. Diagram of DNA “fingerprint” from the Southern blot technique of DNA hybridization to distinguish monoclonal from polyclonal cell populations. The method is as follows: Double-stranded DNA is extracted from lymphocyte suspensions by cell lysis. Restriction endonucleases then are applied to the DNA to cleave it into several fragments. A. The fragments undergo electrophoresis on agarose gel slabs, which separates them based on their size and charge. The fragments then are denatured, which breaks them into single-stranded pieces of DNA. The fragments then are transferred to nitrocellulose filter. The fragments are incubated with 32p-labeled DNA probes, which are single-stranded nucleic acid fragments that hybridize specific base sequences from the original specimen. Probes then are used for regions known to encode for the kappa or gamma light chains, the immunoglobulin heavy chain region, and the T-cell receptor beta chain. Autoradiography is used to identify the radiolabeled material. This yields a blot that varies in intensity, depending on the amount of labeled material present in a given electrophoretic band. Each assessment is performed with a fibroblast control. Fibroblasts express the germline configuration of the light chain, heavy chain, and cell receptor DNA. B. Polyclonal populations express a variety of rearrangements but yield a germline band because it is the most frequent across the population, whereas a monclonal population yields a nongermline band, which represents all of the cells in the population. |