
|
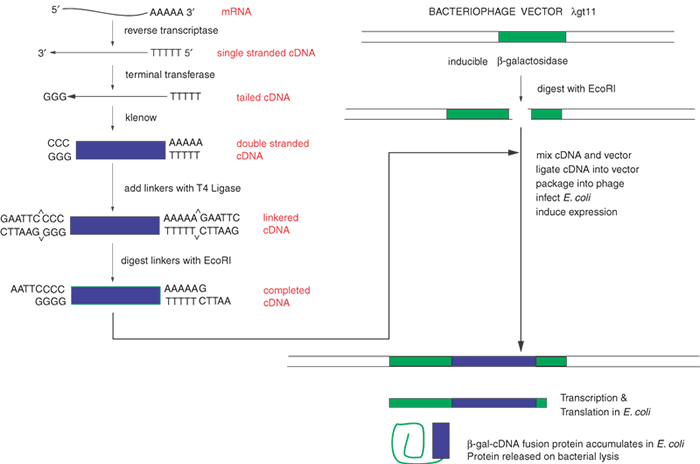
| Fig. 3. Construction of a cDNA Library. To clone a large group of cDNAs from a given tissue, such as the retina, we begin with a few milligrams of RNA. Only approximately 0.5% to 5% of the total RNA represents mRNA; the bulk of the RNA is ribosomal and tRNA. Reverse transcriptase copies the mRNA into single-stranded DNA, complementary DNA (cDNA). This is converted to double-stranded DNA by the Klenow fragment of DNA polymerase I. Linkers are ligated to the ends of the cDNA. When digested with a restriction enzyme, such as EcoRI, these give sticky ends compatible with similarly digested vector. The thousands of different cDNAs are mixed with an excess of the digested vector, and T4 ligase is added. In suitable conditions only one cDNA will be ligated into each vector molecule. The recombinant vector, λgt11 with a cDNA, is packaged to reconstitute viable bacteriophage and can be grown on agar plates seeded with enough Escherichia coli to form a uniform lawn of cells over the surface of the plate. Because of the design of the vector, the infected E. coli will produce some protein derived from the cDNA. This protein also contains some amino acid sequences from the vector, and the protein is a hybrid of bacterial galactosidase and the polypeptide from the cDNA. This is called a fusion protein. The group of bacteriophage resulting from the insertion of the individual cDNAs into the vector molecules is called a cDNA library. |