
|
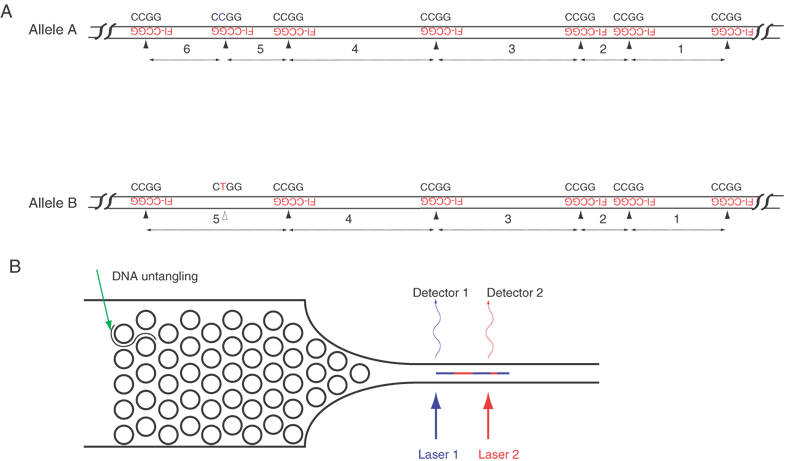
| Fig. 7. Polymorphism detection in a single DNA molecule. Panel A. A fluorescently tagged tetramer (Fl-CCGG in red) is annealed to a genomic DNA fragment. Two different alleles are illustrated, with Allele A having one extra site and Allele B lacking one site. For the sake of clarity, only tetramer binding sites on the upper strand of DNA are illustrated. In comparing two alleles of the same fragment, sequence variations can be detected. Allele B has exactly the same fragment size and order as Allele A for the first 4 fragments, but fragment 5 is larger in Allele B, and Allele A contains two fragments (5 and 6) in place of Allele B's single fragment 5. A critical point is that the former two from Allele A add up to the same size as Allele B's fragment 5. Panel B. The hybridized probe-DNA complex is straightened in the funnel by catching on several pegs. The DNA passes by one or more fluorescence detectors. The DNA molecule moves across the detector at a constant rate of about 1 cm/sec. Because the DNA is straightened and in line with flow, the time elapsed from the detection of one fluorescent probe to the next reflects the number of bases between two adjacent tetramer binding sites. The order and sizes of the fragments can be recorded and compared with the predicted pattern of tetramers from the known human sequence. This identifies the location and length of the fragment. |