
|
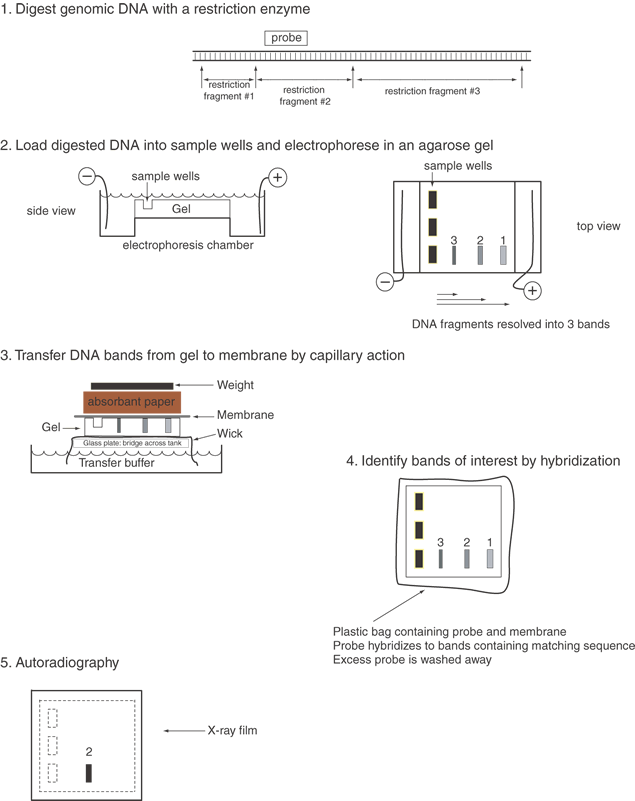
| Fig. 2. Southern blot analysis. To identify a specific genomic DNA fragment among the hundreds of thousands of restriction enzyme fragments in the human genome, we use the Southern blot technique.7 This approach consists of several steps illustrated above. Genomic DNA is prepared and digested with any of several restriction enzymes. The DNA fragments are separated by size on agarose gel electrophoresis. The smaller fragments migrate faster through the agarose fiber meshwork. The fragments of interest can be identified by hybridization with a labeled probe. The DNA bands are first transferred out of the agarose onto a nitrocellulose or nylon membrane by capillary action. The DNA fragments can be permanently attached to the membrane by heat or ultraviolet light treatment. A radiolabeled DNA probe (e.g., a denatured cDNA) in single-stranded form can anneal by base pairing to its counterpart on the membrane. Excess probe DNA is washed off the filter, and by autoradiography of the filter we can visualize the specific DNA band or bands matching the probe. The Southern blot procedure allows us to detect and measure the size of one or a few DNA fragments out of the millions of different genomic DNA fragments produced by digestion with a given restriction enzyme. |